2024
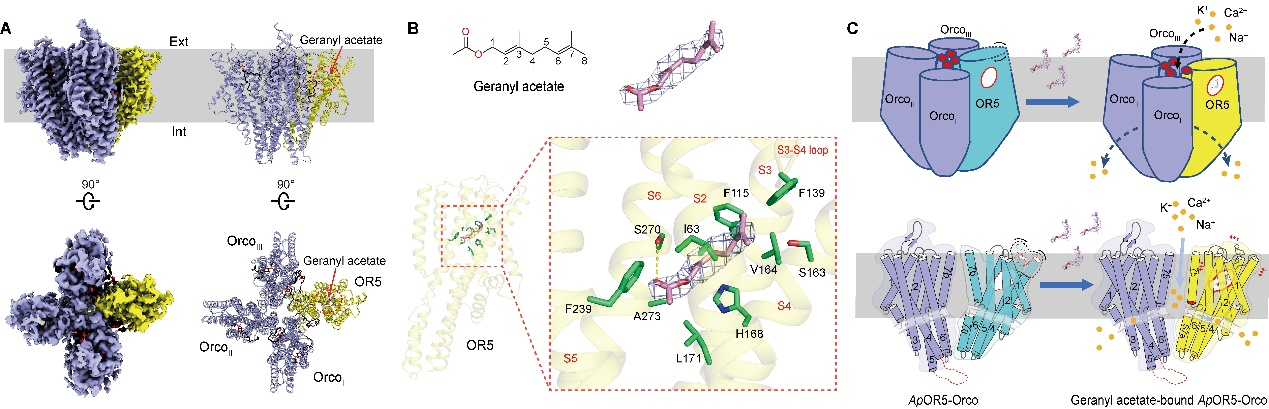
Wang Y#, Qiu L#, Wang B#, Guan Z, Dong Z, Zhang J, Cao S, Yang L, Wang B, Gong Z, Zhang L, Ma W, Liu Z, Zhang D, Wang G* and Yin P*. Structural basis for odorant recognition of the insect odorant receptor OR-Orco heterocomplex. Science. 2024. https://doi.org/10.1126/science.adn6881. https://www.science.org/stoken/author-tokens/ST-1944/full
Wang Q#, Zhuang J#, Huang R#, Guan Z, Yan L, Hong S, Zhang L, Huang C, Liu Z, Yin P*. The architecture of substrate-engaged TOM–TIM23 supercomplex reveals preprotein proximity sites for mitochondrial protein translocation. Cell Discovery. 2024(10)1:19. https://doi.org/10.1038/s41421-023-00643-y. 2024.2.16
2023
Yan X#, Liu F#, Yan J, Hou M, Sun M, Zhang D, Gong Z, Dong X, Tang C*, Yin P*. WTAP–VIRMA counteracts dsDNA binding of the m6A writer METTL3–METTL14 complex and maintains N6-adenosine methylation activity. Cell Discovery. 2023(9)1. doi:10.1038/s41421-023-00604-5. 2023 Oct 3.
Wang J#, Zhou C#, Guan Z,Wang Q, Zhao J, Wang L, Zhang L, Zhang D, Deng X, Ma L*, Yin P*. Plant phytochrome A in the Pr state assembles as an asymmetric dimer. Cell Research. 2023,33(10):802-805. doi.org/10.1038/s41422-023-00847-7. 2023 Oct.
Yan J# , Liu F#, Guan Z, Yan X, Jin X, Wang Q, Wang Z, Yan J, Zhang D, Liu Z, Wu S*, Yin P*. Structural insights into DNA N6 -adenine methylation by the MTA1 complex. Cell Discovery. 2023,9(1):8. doi:10.1038/s41421-022-00516-w. 2023 Jan 20.
Wang L#, Wang Y#, Chang H, Ren H, Wu X, Wen J, Guan Z, Ma L, Qiu L, Yan J, Zhang D, Huang X*, Yin P*. RUP2 facilitates UVR8 redimerization via two interfaces. Plant Communications. 2023(9):100428. doi:10.1016/j.xplc.2022.100428. 2023 Jan 9.
Ma L, Jia H, Shen A, Ding J, Wang X, Wang J, Wan J, Yan J, Zhang D, Dong X*, Yin P*. Two determinants influence CRY2 photobody formation and function. Plant Biotechnology Journal, 2023,21(3):460-462. doi: 10.1111/pbi.13978. 2023 Mar.
2022
Yan X#, Pei K#, Guan Z#, Liu F, Yan J, Jin X, Wang Q, Hou M, Tang C*, Yin P*. AI-empowered integrative structural characterization of m6A methyltransferase complex. Cell Research. 2022,32 (12):1124-1127. doi:10.1038/s41422-022-00741-8. 2022 Dec.
Wang Y#, Wang L#, Guan Z#, Chang H, Ma L, Shen C, Qiu L, Yan J, Zhang D, Li J, Deng X*, Yin P*. Structural insight into UV-B-activated UVR8 bound to COP1. Science Advances. 2022,8(16): eabn3337. doi:10.1126/sciadv.abn3337. 2022 Apr.
2021
Wang Q#, Guan Z#, Qi L#, Zhuang J, Wang C, Hong S, Yan L, Wu Y, Cao X, Cao J, Yan J, Zou T, Liu Z, Zhang D, Yan C, Yin P*. Structural insight into the SAM-mediated assembly of the mitochondrial TOM core complex. Science. 2021,373(6561):1377-1381. doi: 10.1126/science.abh0704. 2021 Aug 26.
Guan Z#, Yan L#, Wang Q, Qi L, Hong S, Gong Z, Yan C*, and Yin P*. Structural insights into assembly of Human Mitochondrial Translocase TOM Complex. Cell Discov. 2021,7(1):22. doi: 10.1038/s41421-021-00252-7. 2021 Apr 13.
Zhan X#, Xue Y #, Guan Z#, Zhou C, Nie Y, Men S, Wang Q, Shen C, Zhang D, Jin S, Tu L*, Yin P*, and Zhang X*. Structural insights into homotrimeric assembly of cellulose synthase CesA7 from Gossypium hirsutum. Plant Biotechnology Journal. 2021,19(8):1579-1587. doi: 10.1111/pbi.13571. 2021 Aug.
Qi L#, Wang Q#, Guan Z#, Wu Y, Shen C, Hong S, Cao J, Zhang X, Yan C* and Yin P*. Cryo-EM structure of the human mitochondrial translocase TIM22 complex. Cell Res. 2021,31(3):369-372.doi: 10.1038/s41422-020-00400-w. 2021 Mar.
2020
Yan J#, Hong S#, Guan Z, He W, Zhang D, and Yin P*. Structural insights into sequence-dependent Holliday junction resolution by the chloroplast resolvase MOC1. Nat Commun. 2020,11(1):1417. doi: 10.1038/s41467-020-15242-8. 2020 Mar 17;
Ma L#, Wang X#, Guan Z Y#, Wang L X, Wang Y D, Zheng L, Gong Z, Shen C C, Wang J, Zhang D L, Liu Z and Yin P*. Structural insights into BIC-mediated inactivation of Arabidopsis cryptochrome 2. Nat. Struct. Mol. Biol. 2020,27(5): 472-479. (Comments by Prof. Chentao Lin, https://www.nature.com/articles/s41594-020-0432-6)
Shen C#, Liu H#, Guan Z, Yan J, Zheng T, Yan W, Wu C, Zhang Q, Yin P*, and Xing Y*. Structural insights into DNA recognition by CCT/NF-YB/YC complex in plant photoperiodic flowering. Plant Cell. 2020 Nov; 32(11):469-3484. DOI: https://doi.org/10.1105/tpc.20.00067
Ma L, Guan Z, Wang Q, Yan X, Wang J, Wang Z, Cao J, Zhang D, Gong X* and Yin P*. Structural insights into the photoactivation of Arabidopsis CRY2. Nat Plants. 2020 Nov 16. doi: 10.1038/s41477-020-00800-1.
2019
Yan J#, Yao Y#, Hong S, Yang Y, Shen C, Zhang Q, Zhang D, Zou T, Yin P*. Delineation of pentatricopeptide repeat codes for target RNA prediction, Nucleic Acids Research. 2019,47(7): 3728-3738. doi: 10.1093/nar/gkz075. 2019 Jan 31.
Huang J#, Dong X#, Gong Z#, Qin L, Yang S, Zhu Y, Wang X, Zhang D, Zou T, Yin P* and Tang C*.Solution structure of the RNA recognition domain of METTL3-METTL14 N6-methyladenosine methyltransferase. Protein&Cell. 2019,10(4):272-284. doi: 10.1007/s13238-018-0518-7. 2019 Apr.
2018
Wang X#, Guan Z, Gong Z, Yan J, Yang G, Liu Y, Yin P*. Crystal structure of WA352 provides insight into cytoplasmic male sterility in rice. BBRC. 2018 May 23; 501(4):898-904. doi.org/10.1016/j.bbrc.2018.05.079.
Huang J#, Yin P*. Structural Insights into N6-methyladenosine (m6A) Modification in the Transcriptome. Genomics, Proteomics & Bioinformatics. 2018 Mar 29; 16(2):85-98. doi: https://doi.org/10.1016/j.gpb.2018.03.001
Ma L, Wang Q, Yuan M, Zou T, Yin P* and Wang S*. Xanthomonas TAL effectors hijack host basal transcription factor IIA α and γ subunits for invasion. BBRC. 2018 Jan 9; 496(2):608-613. doi: 10.1016/j.bbrc.2018.01.059.
Wang Q#, Zhang D#, Guan Z, Li D, Pei K, Liu J, Zou T and Yin P*. DapF stabilizes the substrate-favoring conformation of RppH to stimulate its RNA-pyrophosphohydrolase activity in Escherichia coli. Nucleic Acids Research. 2018 May 28. 46(13): 6880-6892. doi: 10.1093/nar/gky528.
Liu J, Guan Z, Liu H, Qi L, Zhang D, Zou T, and Yin P*. Structural insights into the substrate recognition mechanism of Arabidopsis GPP-bound NUDX1 for noncanonical monoterpene biosynthesis. Mol. Plant. 2018,11(1):218-221. doi:10.1016/j.molp.2017.10.006. 2018 Jan 8.
Yan J#, Zhang Q and Yin P*. RNA editing machinery in plant organelles. Sci China Life Sci. 2018,61(2):162-169. doi: 10.1007/s11427-017-9170-3. 2018 Feb.
2017
Yan J#, Zhang Q#, Guan Z, Wang Q, Li L, Ruan F, Lin R, Zou T, Yin P*. MORF9 increases the RNA-binding activity of PLS-type pentatricopeptide repeat protein in plastid RNA editing. Nature Plants. 2017 Apr 10; 3:17037. doi: 10.1038/nplants.2017.37.
Wang X, Huang J, Zou T, Yin P*. Human m6A writers: Two subunits, Two roles. RNA Biol. 2017 Mar 4;14(3):300-304. doi: 10.1080/15476286.2017.1282025. Epub 2017 Jan 25.
Zhao W#, Zhou C#, Guan Z, Yin P*, Chen F* and Tang Y*. Structural Insights into the Inhibition of Tubulin by the Antitumor Agent 4β-(1,2,4-triazol-3-ylthio)-4-deoxypodophyllotoxin. ACS Chem. Biol. 2017,12(3):746–752. doi:10.1021/acschembio.6b00842. 2017 Mar 17.
2016
Zhang D#, Liu Y#, Wang Q, Guan Z, Wang J, Liu J, Zou T, Yin P*. Structural basis of prokaryotic NAD-RNA decapping by NudC. Cell Research. 2016 Sep;26(9):1062-6. doi: 10.1038/cr.2016.98. Epub 2016 Aug 26.
Wang X#, Feng J#, Xue Y#, Guan Z, Zhang D, Liu Z, Gong Z, Wang Q, Huang J, Tang C, Zou T, Yin P*. Structural basis of N6-adenosine methylation by the METTL3-METTL14 complex. Nature. 2016,534(7608):575-8. doi: 10.1038/nature18298. 2016 Jun 23.
Shen C#, Zhang D#, Guan Z#, Liu Y, Yang Z, Yang Y, Wang X, Wang Q, Zhang Q, Fan S, Zou T, Yin P*. Structural basis for the specific single-stranded RNA recognition by designer pentatricopeptide repeat protein. Nat. Commun. 2016,7:11285.doi:10.1038/ncomms11285. 2016 Apr 18.
2015
Shen C#, Wang X#, Liu Y, Li Q, Yang Z, Yan N, Zou T, Yin P*. Specific RNA recognition by designer pentatricopeptide repeat protein. Mol Plant. 2015,8(4):667-70. doi: 10.1016/j.molp.2015.01.001. 2015 Apr;
2014
Li Q, Yan C, Xu H, Wang Z, Long J, Li W, Wu J, Yin P*, Yan N*. Examination of the dimerization states of the single-stranded RNA-recognition protein PPR10. J Biol Chem. 2014 Nov 7; 289 (45):31503-12. doi: 10.1074/jbc.M114.575472.
He Y#, Hao Q, Li W, Yan C, Yan N*, Yin P*. Identification and characterization of ABA receptors in Oryza sativa. PloS One. 2014,9(4): e95246. doi:10.1371/journal.pone.0095246 2014 Apr 17;
2013
Yin P#, Li Q#, Yan C, Liu Y, Liu J, Yu F, Wang Z, Long J, He J, Wang H, Wang J, Zhu J, Shi Y, Yan N*. Structural basis for the modular recognition of single-stranded RNA by PPR proteins. Nature. 2013,504 (7478): 168-171. doi: 10.1038/nature12651. 2013 Dec 5;


 旧版首页
旧版首页 全新首页
全新首页
 NCS
NCS  学术网站链接
学术网站链接  蛋白质化学工具
蛋白质化学工具  蛋白质平台
蛋白质平台  实用工具网站
实用工具网站 
 学术动态
学术动态  人文建设
人文建设  学术交流
学术交流  人物随感
人物随感  其他新闻
其他新闻  学术周边
学术周边  科学大事件
科学大事件 
 Online PDB
Online PDB  2024年
2024年  2023年
2023年  2022年
2022年  2021年
2021年  2020年
2020年  2019年
2019年  2018年
2018年  2017年
2017年  2016年
2016年  2015年
2015年  2014年
2014年  Prior to 2014
Prior to 2014 
 实验室概况
实验室概况  研究方向
研究方向 
 Thesis Defence
Thesis Defence  Lab Meeting
Lab Meeting  Fun Time
Fun Time  Awards
Awards  Talk
Talk  Notice
Notice  Experiment notebook
Experiment notebook  实验室安全教育
实验室安全教育 
 Groupleader
Groupleader  Associate Professor
Associate Professor  Postdoctoral Fellows
Postdoctoral Fellows  Ph.D Candidates
Ph.D Candidates  Master Candidates
Master Candidates  Undergraduate
Undergraduate  Collaborator
Collaborator  Former Lab Members
Former Lab Members 

 鄂公网安备 42011102000808号
鄂公网安备 42011102000808号 
